Linear Mixed Effects Models in R
An introduction for linguistic students
Chenzi Xu
University of Oxford
2021/12/12 (updated: 2022-02-18)
chenzi.xu@rbind.io
Did you do the pre-workshop setup?
https://chenzixu.rbind.io/talk/lme/
We assume:
You have basic knowledge about R and R studio.
You have basic inferential statistical knowledge.
You want to know more about linear mixed models
high school arithmetic and algebra. Elementary concepts of probability theory are assumed; the sum and the product rules, and the fact that the probabilities of all possible events must sum to 1, are the extent of the knowledge assumed.
Outline
Inferential Stats Recap
Linear Regression
Linear Mixed Effects Models
Shrinkage in linear mixed models
Report Writing
Why LME model?
Inferential Stats Recap
Inferential Stats Recap
Central limit theorem (magic)
Hypothesis testing: the (humble) t-test
Central limit theorem (magic)
Definition and Significance
The picture is from Wikipedia.
The sampling distribution of means is normal, provided two conditions are met: (a) the sample size should be large enough (b) μ and σ are defined for the probability density or mass function that generated the data.
We can derive an estimate of the distribution of (hypothetical) sample means from a single sample of n independent data points
CLT is NOT the distribution that generated the data, but the sampling distribution of the sample means under repeated sampling.

SE estimates how much error there is, across repeated sampling, in our estimate of the population mean using sample mean. Our estimation will be more precise for larger sample size or lower variability.
Hypothesis testing: the (humble) t-test
NHST procedure
- Define the null hypothesis μ0
- Given data of size n, estimate ˉy, standard deviation s, estimate the standard error SE=s/√n.
- The distance between the sample mean and the hypothesized mean can be written in SE units as follows.t×SE=ˉy−μ Thus the sample mean is t standard errors away from the hypothesized mean: t=ˉy−μ0s/√n
- Reject null hypothesis if the observed t-value is “large”.
- Fail to reject the null hypothesis, if the observed t-value is “small”.
One of the simplest statistical tests that one can do with continuous data: the t-test. the logic of these more complex tests will essentially be more of the same
uncertainty in the estimates of the values as the scale on which distances are measured.
Hypothesis testing: the (humble) t-test
Under repeated sampling, the observed t-value is seen as an instance from a random variable T.
T∼t(n−1)
The PDF of T, the t-distribution, is defined in terms of of n with n-1 degrees of freedom.
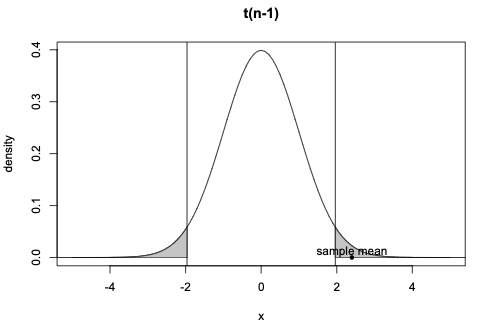
Critical t-value: the t-value such that P(T>|t|)=0.05
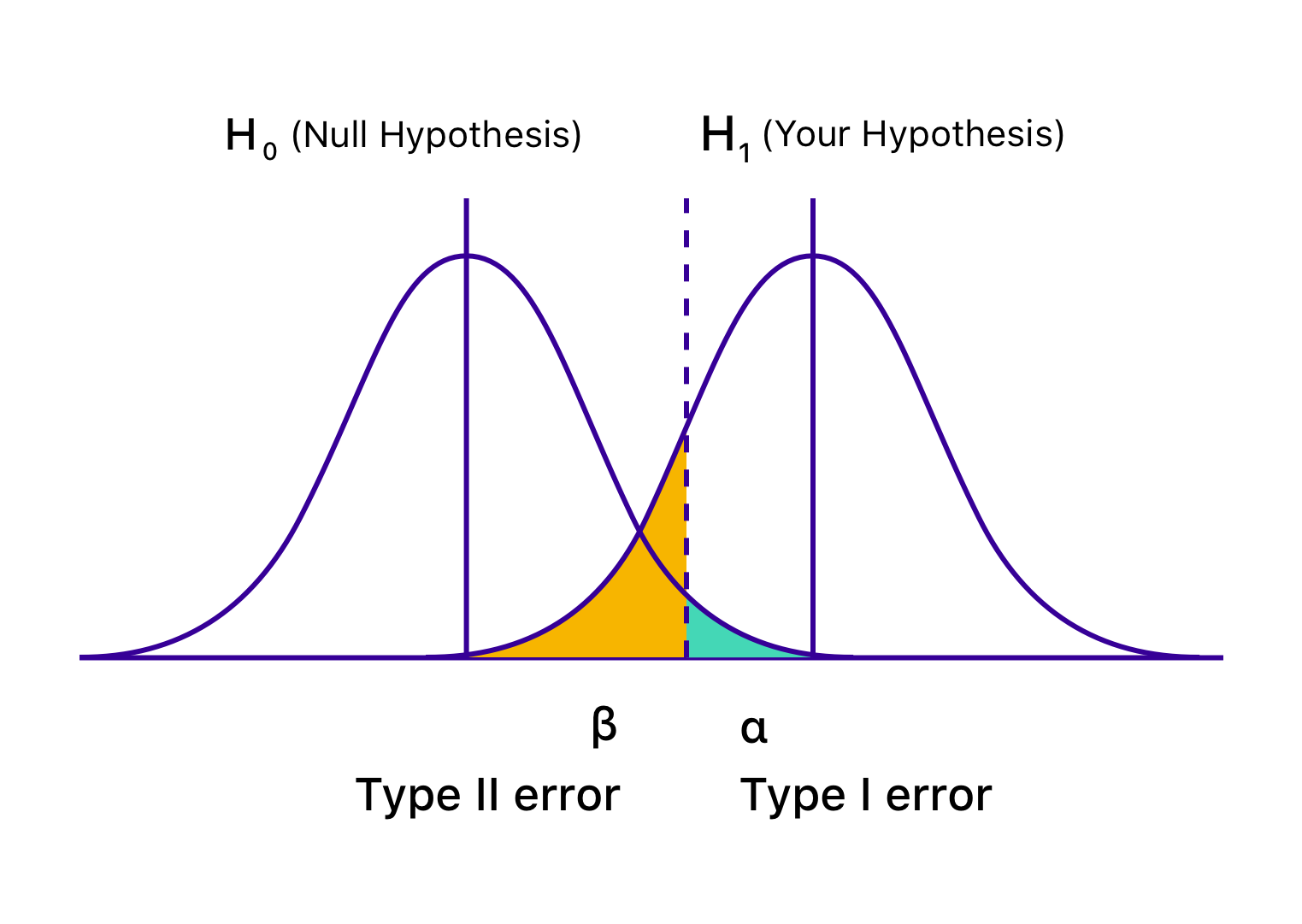
Type I error: Reject the null when it’s true
Type II error: Fail to reject the null when it’s false
p-value: the probability of obtaining the observed t-value, or some value more extreme than that, under the assumption that the null is true.
The p-value is always interpreted with reference to the pre-defined Type I error. Conventionally, we reject the null if p<0.05
Two possible decisions: reject the null or fail to reject the null. Two possible worlds: the null is true or false. The first picture is from Vasishth's book; The second picture is from https://vwo.com/blog/errors-in-ab-testing/
The t-test is just a special, limited example of a general linear model. ANOVA is also a special, limited version of the linear model.(categorical predictors. more than two levels.) general linear model: A mathematical model comparing how one or more independent variables affect a continuous dependent variable.
df measures number of pieces of independent information that go into estimateing some parameters.
p-values are NOT the probability the null hypothesis is true, and does not measure strength of an effect.
Linear Regression
Linear Regression
Formal forms
A (too) simple case study
Understanding output
Assumptions check
Formal forms
A simple example of linear model is Yi=β0+β1Xi+ei Where Yi is the measured value of the dependent variable for observation i; Xi is the explanatory variable or predictor; Yi is modelled in terms of an intercept β0 plus the effect of predictor Xi weighted by coefficient β1 and error term ei.
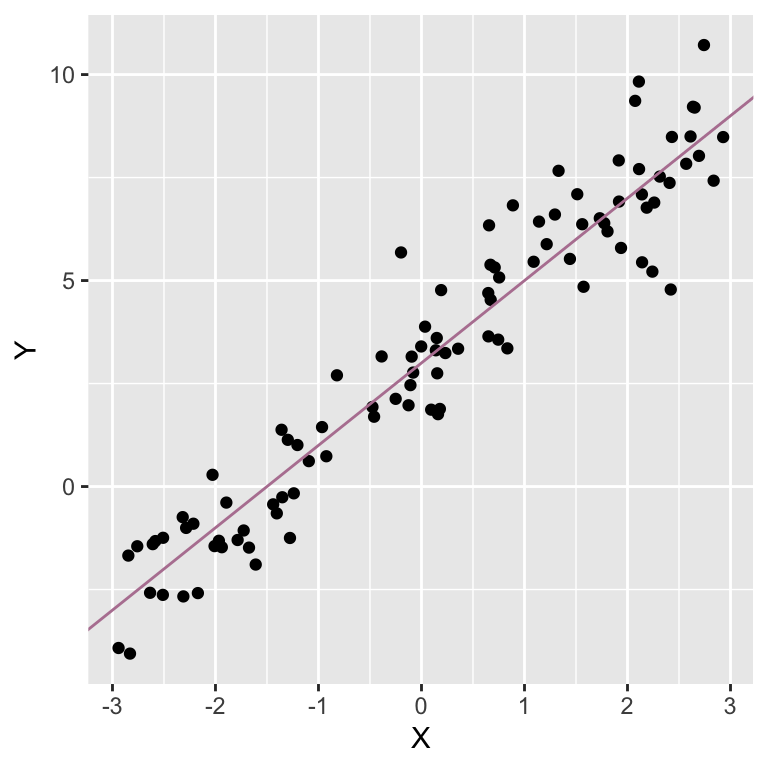
Simulated data illustrating linear model Yi=3+2Xi+ei. The most common method for fitting a regression line is least-squares.
A linear model is a model that is linear in the coefficients; each coefficient is only allowed to be set to the power of one.
Non-linear relationships can be analysed using a linear model.
LME an extension of the General Linear Model that underlies simpler approaches such as ANOVA, t-test, and classical regression. Many people assume that 'linear models' can only capture linear relationships, i.e., relationships that can be described by a straight line (or a plane). This is false. A linear model is a weighted sum of various terms, and each term has a single predictor (or a constant) that is multiplied by a coefficient.
A simple case study
Research question
Assume you were interested in whether the voice pitch of males and females differs, and if so, by how much.
Data
pitch = c(233,204,242,130,112,142)sex = c(rep("female",3), rep("male",3))my.df = data.frame(sex,pitch)my.df# sex pitch# 1 female 233# 2 female 204# 3 female 242# 4 male 130# 5 male 112# 6 male 142Formula
Relationship of interest pitch∼sex
"Pitch predicted by / as a function of sex" pitch∼sex+ϵ ϵ stands for all other things that affect pitch, random or controllable in our experiment. Structural or systematic + random or probabilistic
model = lm(pitch ~ sex, my.df)contrasts(as.factor(my.df$sex))## male## female 0## male 1Dummy coding for categorical factors
This example is taken from Bodo Winter's tutorial.
Dummy coding for categorical factors contrasts(as.factor(my.df$sex))
Understanding output
summary(model)# # Call:# lm(formula = pitch ~ sex, data = my.df)# # Residuals:# 1 2 3 4 5 6 # 6.667 -22.333 15.667 2.000 -16.000 14.000 # # Coefficients:# Estimate Std. Error t value Pr(>|t|) # (Intercept) 226.33 10.18 22.224 2.43e-05 ***# sexmale -98.33 14.40 -6.827 0.00241 ** # ---# Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1# # Residual standard error: 17.64 on 4 degrees of freedom# Multiple R-squared: 0.921, Adjusted R-squared: 0.9012 # F-statistic: 46.61 on 1 and 4 DF, p-value: 0.002407R2: "variance accounted for or explained by"
Multiple R-Squared: how much variance in our data is accounted for by our model
Adjusted R-Squared: how much variance is “explained” and how many fixed effects are used
p-value: The probability of our data, given that our null hypothesis "sex has no effect on pitch" is true.
Reporting:
We constructed a linear model of pitch as a function of sex. This model was significant (F(1,4)=46.61,p<0.01).
Understanding output
summary(model)# # Call:# lm(formula = pitch ~ sex, data = my.df)# # Residuals:# 1 2 3 4 5 6 # 6.667 -22.333 15.667 2.000 -16.000 14.000 # # Coefficients:# Estimate Std. Error t value Pr(>|t|) # (Intercept) 226.33 10.18 22.224 2.43e-05 ***# sexmale -98.33 14.40 -6.827 0.00241 ** # ---# Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1# # Residual standard error: 17.64 on 4 degrees of freedom# Multiple R-squared: 0.921, Adjusted R-squared: 0.9012 # F-statistic: 46.61 on 1 and 4 DF, p-value: 0.002407mean(my.df[my.df$sex=="female",]$pitch)# [1] 226.3333mean(my.df[my.df$sex=="male",]$pitch)# [1] 128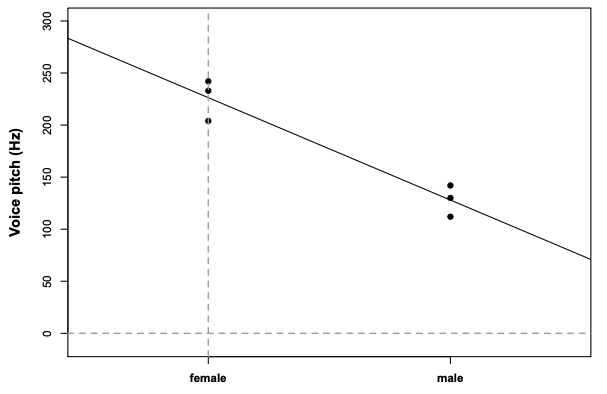
Assumptions check
(1) Linearity
The fitted values (line) are the predicted means, and residuals are the vertical deviations from this line.
When the residual plot shows a curvy or non-linear patterns, it indicates a violation of the linearity assumption.
plot(fitted(model),residuals(model))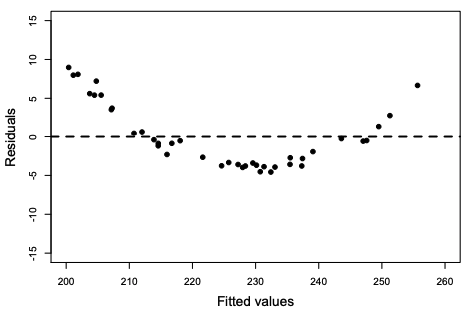
What to do if your residual plot indicates nonlinearity?
- Reconsider the fixed effects
- Perform non-linear transformation of your variables
- Turn to different class of models such as logistic models
conditions that have to be satisfied in order for the linear model to be meaningful.
Assumptions check
(2) Absence of collinearity
When two predictors are correlated with each other, they are collinear.
Preempt the problem in the design stage
Use dimension reduction techniques such as Principal Component Analysis that transform correlated variables into a smaller set of variables
(3) Homoskedasticity
The variance of your data should be approximately equal across the range of the predicted values.
- Transform your data
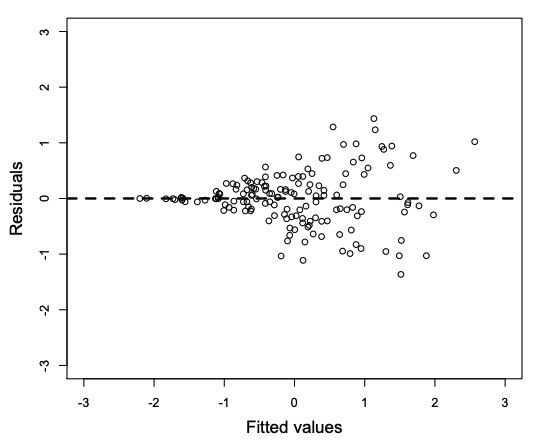
Assumptions check
(4) Absence of influential data points
dfbeta(model)# (Intercept) sexmale# 1 3.333333e+00 -3.333333# 2 -1.116667e+01 11.166667# 3 7.833333e+00 -7.833333# 4 -1.359740e-16 1.000000# 5 1.087792e-15 -8.000000# 6 -9.518180e-16 7.000000DFbeta values: the coefficients to be adjusted if a particular data point is excluded (“leave-one-out diagnostics”).
Eyeball the DFbeta values: any sign changed? different by half of the absolute value?
Run the analysis with influential data and then again without them. Compare and discuss the results.
dfbeta() for linear models doesn’t work for mixed models. You can check out the package influence.ME (Nieuwenhuis, te Grotenhuis, & Pelzer, 2012), or program a for loop that does the leave-one-out diagnostics by hand.
Assumptions check
(5) Normality of residuals
- a histogram or a Q-Q plot of the residuals
hist(residuals(model))qqnorm(residuals(model))(6) Independence
- Preempt the problem in the design stage
- Resolve non-independencies at the analysis stage. → Mixed models
Linear Mixed Effects Model
Linear Mixed Effects Model
Data
Model
Assumption
Understanding the output
Model selection
Data
Research Question
We're interested in the relationship between pitch and politeness (Winter & Grawunder, 2012).
- Politeness: formal/polite and informal register (categorical factor)
- multiple measures per subject (inter-dependent!)
data = read.csv("http://www.bodowinter.com/tutorial/politeness_data.csv")head(data)# subject gender scenario attitude frequency# 1 F1 F 1 pol 213.3# 2 F1 F 1 inf 204.5# 3 F1 F 2 pol 285.1# 4 F1 F 2 inf 259.7# 5 F1 F 3 pol 203.9# 6 F1 F 3 inf 286.9which(!complete.cases(data))# [1] 39a few missing values provide no problems for our mixed model analyses
Model
Linear regression pitch∼politeness+ϵ Multiple regression (fixed effects only) pitch∼politeness+sex+ϵ Add a random effect for subject: assume a different "baseline" pitch value for each subject. We can assume different random intercepts for each subject. pitch∼politeness+sex+(1|subject)+ϵ
boxplot(frequency~subject, data)Crossed random effects for subjects and for items. Different scenarios for formal and informal speech (by-item variation) pitch∼politeness+sex+(1|subject)+(1|item)+ϵ
Model in R
library(lme4)data = data %>% mutate(attitude=as.factor(attitude), gender=as.factor(gender), subject=as.factor(subject))politeness.model0 = lmer(frequency ~ attitude + (1|subject) + (1|scenario), data=data)politeness.model = lmer(frequency ~ attitude + gender + (1|subject) + (1|scenario), data=data)summary(politeness.model)# Linear mixed model fit by REML. t-tests use Satterthwaite's method ['lmerModLmerTest']# Formula: frequency ~ attitude + gender + (1 | subject) + (1 | scenario)# Data: data# # REML criterion at convergence: 775.5# # Scaled residuals: # Min 1Q Median 3Q Max # -2.2591 -0.6236 -0.0772 0.5388 3.4795 # # Random effects:# Groups Name Variance Std.Dev.# scenario (Intercept) 219.5 14.81 # subject (Intercept) 615.6 24.81 # Residual 645.9 25.41 # Number of obs: 83, groups: scenario, 7; subject, 6# # Fixed effects:# Estimate Std. Error df t value Pr(>|t|) # (Intercept) 256.846 16.116 5.432 15.938 9.06e-06 ***# attitudepol -19.721 5.584 70.054 -3.532 0.000735 ***# genderM -108.516 21.013 4.007 -5.164 0.006647 ** # ---# Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1# # Correlation of Fixed Effects:# (Intr) atttdp# attitudepol -0.173 # genderM -0.652 0.004Understanding the output
politeness.model0 (top) politeness.model (bottom)
Scenario (item) has much less variability than subject.
Residual: The “random” deviations from the predicted values that are not due to subjects and items, ϵ.
The variation that’s associated with the random effect “subject” dropped considerably → gender was confounded with the variation that is due to subject.
Random effects
Random effects: Groups Name Variance Std.Dev. scenario (Intercept) 219 14.80 subject (Intercept) 4015 63.36 Residual 646 25.42 Number of obs: 83, groups: scenario, 7; subject, 6Random effects: Groups Name Variance Std.Dev. scenario (Intercept) 219.5 14.81 subject (Intercept) 615.6 24.81 Residual 645.9 25.41 Number of obs: 83, groups: scenario, 7; subject, 6shifted a considerable amount of the variance that was previously in the random effects component(differences between male and female individuals) to the fixed effects component
Understanding the output
politeness.model0 (top) politeness.model (bottom)
attitudepol is the slope for the categorical effect of politeness.
Pitch is lower in polite speech than in informal speech, by about 20 Hz.
lmer() took whatever comes first in the alphabet to be the reference level.
The second Intercept represents the female for the informal condition.
Fixed effects
Fixed effects: Estimate Std. Error t value (Intercept) 202.588 26.754 7.572 attitudepol -19.695 5.585 -3.527Fixed effects: Estimate Std. Error t value (Intercept) 256.846 16.116 15.938 attitudepol -19.721 5.584 -3.532 genderM -108.516 21.013 -5.164Understanding the output
Coding schemes
A categorical variable of n levels is entered into a regression analysis as a sequence of n−1 variables. Generally or by default in R, we use dummy coding for them.
contrasts(data$attitude)# pol# inf 0# pol 1# change the reference levelcontrasts(data$attitude) = contr.treatment(2, base = 2)contrasts(data$attitude)# 1# inf 1# pol 0There are also other coding schemes such as effects coding including contrast coding, sum coding and Helmert coding. Different coding system may help answer different research questions. It also greatly changes the way you interpret your output.
Understanding the output
Statistical significance
Unfortunately, p-values for mixed models aren’t as straightforward as they are for the linear model. There are multiple approaches, and there’s a discussion surrounding these, with sometimes wildly differing opinions about which approach is the best.
There is a package lmerTest that provides p values in type I, II or III anova and summery tables for lmer model fits via Satterthwaite's degrees of freedom method or Kenward-Roger method.
library(lmerTest)politeness.model = lmer(frequency ~ attitude + gender + (1|subject) + (1|scenario), data=data)anova(politeness.model)# Type III Analysis of Variance Table with Satterthwaite's method# Sum Sq Mean Sq NumDF DenDF F value Pr(>F) # attitude 8056.2 8056.2 1 70.054 12.473 0.0007351 ***# gender 17225.2 17225.2 1 4.007 26.669 0.0066467 ** # ---# Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1The F-value is actually the quotient of the ratio: effect variance/error variance; between-group variance/within-group variance. All of this means that the larger an F-value, the better for you to find a “significant” effect, a consistent pattern that is unlikely due to chance. Now, basically, what we’re doing with an ANOVA is the following: we look at how unexpected an F-value that we obtained in our study is. A very large F-value means that the between-group variance (the effect variance) exceeds the within-group variance (the error variance) by a substantial amount. The p-value then just gives a number to how likely a particular F-value is going to occur, with lower p-values indicating that the probability of obtaining that particular F-value is pretty low.
Understanding the output
Statistical significance
Likelihood Ratio Test as a means to attain p values. Likelihood is the probability of seeing the data you collected given your model.
politeness.null = lmer(frequency ~ gender + (1|subject) + (1|scenario), data=data, REML=FALSE)politeness.full = lmer(frequency ~ attitude + gender + (1|subject) + (1|scenario), data=data, REML=FALSE)anova(politeness.null, politeness.full)# Data: data# Models:# politeness.null: frequency ~ gender + (1 | subject) + (1 | scenario)# politeness.full: frequency ~ attitude + gender + (1 | subject) + (1 | scenario)# npar AIC BIC logLik deviance Chisq Df Pr(>Chisq) # politeness.null 5 816.72 828.81 -403.36 806.72 # politeness.full 6 807.10 821.61 -397.55 795.10 11.618 1 0.0006532 ***# ---# Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1“… politeness affected pitch ( χ2(1)=11.62,p=0.00065 ), lowering it by about 19.7Hz±5.6 (standard errors)...”
Model selection
coef(politeness.model)# $scenario# (Intercept) attitude1 genderM# 1 223.6187 19.72111 -108.5163# 2 243.7081 19.72111 -108.5163# 3 248.5330 19.72111 -108.5163# 4 257.7546 19.72111 -108.5163# 5 235.1891 19.72111 -108.5163# 6 224.9513 19.72111 -108.5163# 7 226.1215 19.72111 -108.5163# # $subject# (Intercept) attitude1 genderM# F1 223.2175 19.72111 -108.5163# F2 247.5443 19.72111 -108.5163# F3 240.6137 19.72111 -108.5163# M3 265.5072 19.72111 -108.5163# M4 242.5036 19.72111 -108.5163# M7 203.3646 19.72111 -108.5163# # attr(,"class")# [1] "coef.mer"Random intercept model
We accounted for baseline-differences in pitch, but the effect of politeness is the same for all subjects and items.
What if the effect of politeness might be different for different subjects?
Model selection
Random slope model
politeness.model1 = lmer(frequency~attitude + gender + (1+attitude|subject) + (1+attitude|scenario), data = data)coef(politeness.model1)Random intercept model
$subject (Intercept) attitude2 genderMF1 242.9386 -19.72111 -108.5163F2 267.2654 -19.72111 -108.5163F3 260.3348 -19.72111 -108.5163M3 285.2283 -19.72111 -108.5163M4 262.2248 -19.72111 -108.5163M7 223.0858 -19.72111 -108.5163Random slope model (correlated)
$subject (Intercept) attitude2 genderMF1 243.2804 -20.49940 -111.1058F2 267.1173 -19.30447 -111.1058F3 260.2849 -19.64697 -111.1058M3 287.1024 -18.30263 -111.1058M4 264.6698 -19.42716 -111.1058M7 226.3911 -21.34605 -111.1058Despite individual variation, there is also consistency in how politeness affects the voice: for all of our speakers, the voice tends to go down when speaking politely, but for some people it goes down slightly more so than for others.
The effect of politeness (attitudepol) is different for each subject and item. It’s always negative and many of the values are quite similar to each
other.
Model selection
Interaction terms
An interaction, attitude:sex, occurs when one variable changes how another variable affects the response variable. Alternative notation: attitude*sex = attitude + sex + attitude:sex
Correlation between intercepts and slopes
Using || in (1 + attitude || subject) for example, you are forcing the correlation between the random slopes and the intercepts to be zero.
Overfitting
overfitting occurs when your model has too small a sample size and too many predictor variables. You may get a warning / error that your model wouldn’t converge.
Model selection
How to choose your model?
Depend on your research question and data structure
Exploratory VS confirmatory context
Design variables (theoretical interests) VS control variables
Data-driven (e.g. improve the model fit) VS Design-driven
Top-down VS Bottom-up
Some prefer to proceed incrementally, taking time to examine data.
One should always be cautious of creating a model that is too complex.
Some researchers have shown via simulations that mixed models without random slopes have a relatively high Type I error rate (Schielzeth & Forstmeier, 2009; Barr, Levy, Scheepers, & Tilly, 2013)
Barr et al. (2013) recommend that you should “keep it maximal” with respect to your random effects structure, at least for controlled experiments.
We were not interested in gender differences, but they are well worth controlling for. This is why we had random slopes for the effect of attitude (by subjects and item) but not gender.
If you have predictors that represent control variables, over which you do not intend to perform statistical tests, it is unlikely that random slopes are needed.
Model selection
Limitations
"Singular fit" warning or "not converging" error
- We need to simplify our model and remove the random slopes or insignificant terms
(
step()does it automatically for you).
But what if we believe there might be some merit to having subject-specific slopes?
Bayes to the rescue
Bayesian mixed effects method are powerful alternatives to more commonly used Frequentist approaches.
There are two major approaches to fit the model parameters. The principle of maximum likelihood: pick parameter values that maximize the probability of your data Y.
Bayesian inference: put a probability distribution on the model parameters and update it on the basis of what parameters best explain the data.
Shrinkage in linear mixed models
Shrinkage in linear mixed models
Data
Complete pooling
No pooling
Partial pooling
Data
Research question
The effects of sleep deprivation on psychomotor performance / reaction time (Belenky et al., 2003)
Multilevel data with continuous predictor
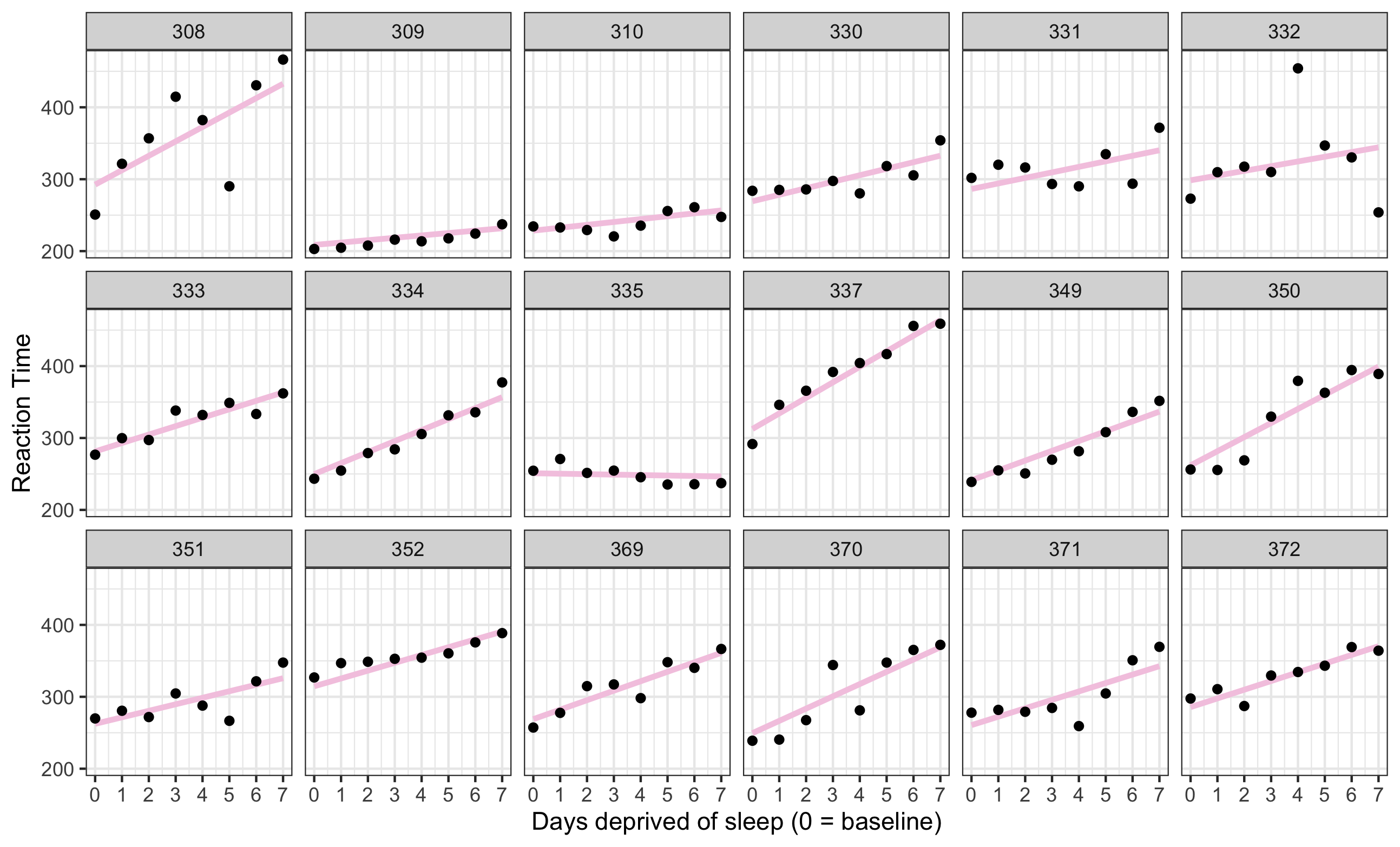
Plot the data
sleep2 <- sleepstudy %>% filter(Days >= 2) %>% mutate(days_deprived = Days - 2)ggplot(sleep2, aes(x = days_deprived, y = Reaction)) + geom_point() + scale_x_continuous(breaks = 0:7) + facet_wrap(~Subject) + labs(y = "Reaction Time", x = "Days deprived of sleep (0 = baseline)")Documentation for the dataset ?sleepstudy
Complete pooling
One-size-fit-all: this approach estimates a single intercept and slope for the entire dataset, ignoring the fact that different subjects might vary in their intercepts or slopes.
It assumes that all observations are independent.
RT=β0+β1Day+e
cp_model <- lm(Reaction ~ days_deprived, sleep2)summary(cp_model)ggplot(sleep2, aes(x = days_deprived, y = Reaction)) + geom_abline(intercept = coef(cp_model)[1], slope = coef(cp_model)[2], color = '#f4cae2', size = 1.5) + geom_point() + scale_x_continuous(breaks = 0:7) + facet_wrap(~Subject, nrow = 3) + labs(y = "Reaction Time", x = "Days deprived of sleep (0 = baseline)")Complete pooling
One-size-fit-all: this approach estimates a single intercept and slope for the entire dataset, ignoring the fact that different subjects might vary in their intercepts or slopes.
It assumes that all observations are independent.
RT=β0+β1Day+e
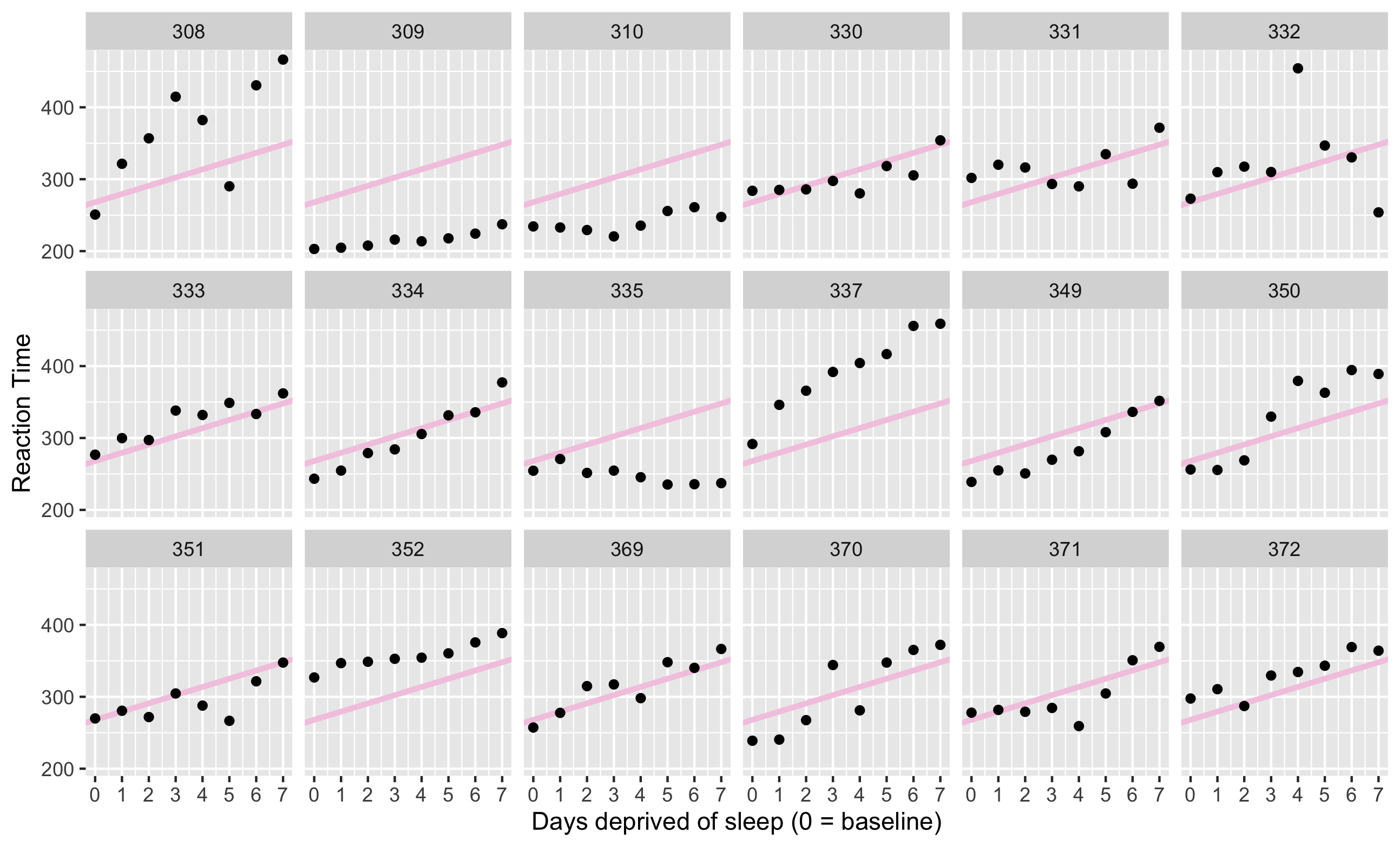

No pooling
This approach fits separate lines for each participant. The estimate for each participant will be completely uninformed by the estimates for the other participants.
There is no overall population intercept and slope that is being estimated.
(1) Run separate regressions for each participant
(2) Run multiple regression
sleep2 %>% pull(Subject) %>% is.factor()np_model <- lm(Reaction ~ days_deprived + Subject + days_deprived:Subject, data = sleep2) summary(np_model)all_intercepts <- c(coef(np_model)["(Intercept)"], coef(np_model)[3:19] + coef(np_model)["(Intercept)"])all_slopes <- c(coef(np_model)["days_deprived"], coef(np_model)[20:36] + coef(np_model)["days_deprived"])ids <- sleep2 %>% pull(Subject) %>% levels() %>% factor()np_coef <- tibble(Subject = ids, intercept = all_intercepts, slope = all_slopes)No pooling
multiple regression
This approach fits separate lines for each participant. The estimate for each participant will be completely uninformed by the estimates for the other participants.
There is no overall population intercept and slope that is being estimated.
ggplot(sleep2, aes(x = days_deprived, y = Reaction)) + geom_abline(data = np_coef, mapping = aes(intercept = intercept, slope = slope), color = '#f4cae2', size = 1.5) + geom_point() + theme_bw() + scale_x_continuous(breaks = 0:7) + facet_wrap(~Subject, nrow=3) + labs(y = "Reaction Time", x = "Days deprived of sleep (0 = baseline)")No pooling
multiple regression
This approach fits separate lines for each participant. The estimate for each participant will be completely uninformed by the estimates for the other participants.
There is no overall population intercept and slope that is being estimated.
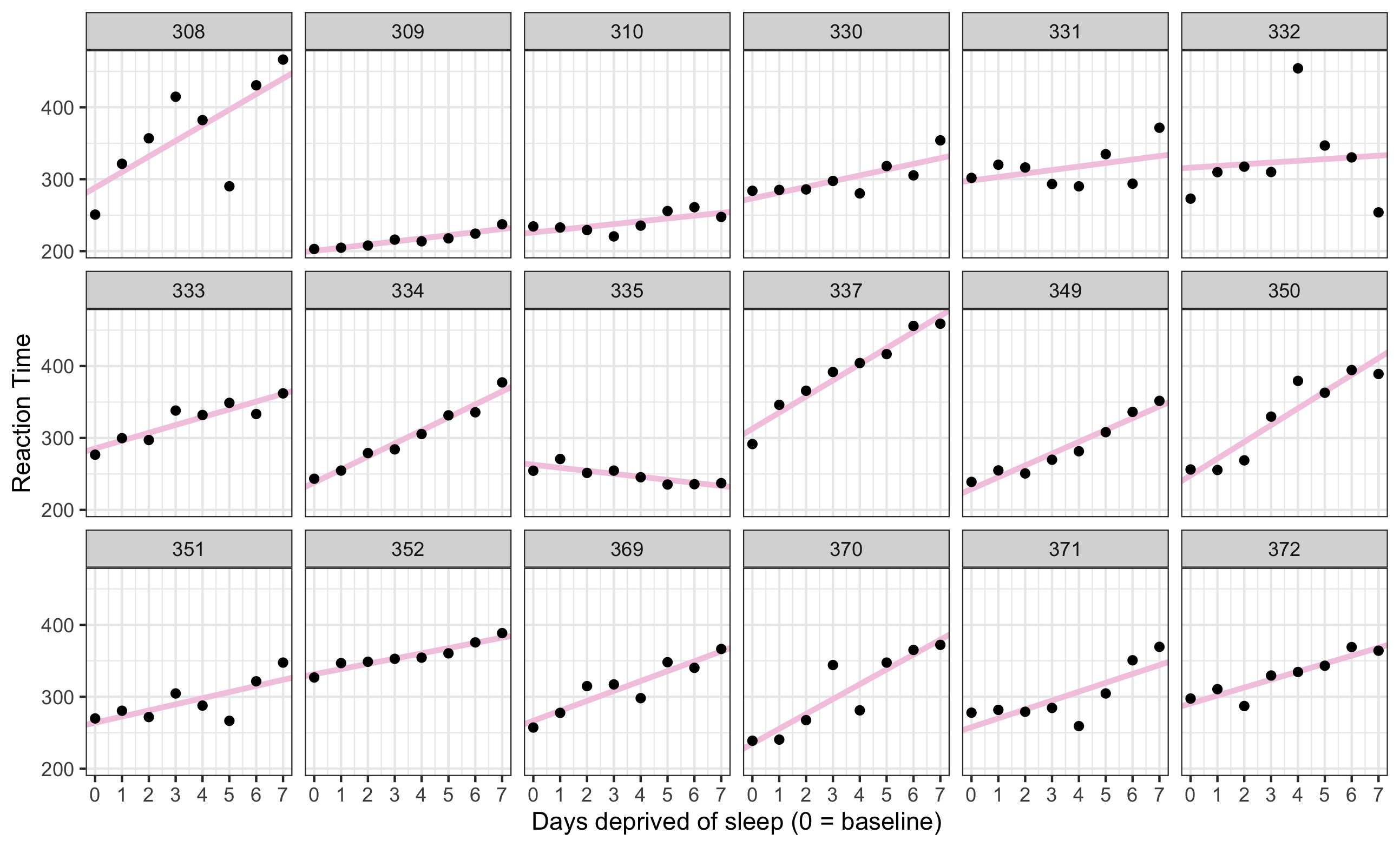
No pooling
multiple regression
This approach fits separate lines for each participant. The estimate for each participant will be completely uninformed by the estimates for the other participants.
There is no overall population intercept and slope that is being estimated.
A one-sample t-test:
np_coef %>% pull(slope) %>% t.test()# # One Sample t-test# # data: .# t = 6.1971, df = 17, p-value = 9.749e-06# alternative hypothesis: true mean is not equal to 0# 95 percent confidence interval:# 7.542244 15.328613# sample estimates:# mean of x # 11.43543The mean slope of 11.435 is significantly different from zero, t(17)=6.197,p<.001.
Partial pooling
It would be desirable to improve our estimates for individual participants by taking advantage of what we know about the other participants.
It would improve generalization to the population.
We are interested in the 18 subjects as examples drawn from a larger population of potential subjects.
LME model estimates values for the population, and pulls the estimates for individual subjects toward those values, a statistical phenomenon known as shrinkage.
Level 1 (for subject i):
RTi=β0i+β1iDayi+ei
Level 2 (for subject i):
β0i=γ0+Subject0i β1i=γ1+Subject1i
Partial pooling happens when you treat a factor as a random instead of fixed in your analysis.
Partial pooling
It would be desirable to improve our estimates for individual participants by taking advantage of what we know about the other participants.
It would improve generalization to the population.
We are interested in the 18 subjects as examples drawn from a larger population of potential subjects.
pp_mod <- lmer(Reaction ~ days_deprived + (days_deprived | Subject), sleep2)summary(pp_mod)newdata <- crossing( Subject = sleep2 %>% pull(Subject) %>% levels() %>% factor(), days_deprived = 0:7)newdata2 <- newdata %>% mutate(Reaction = predict(pp_mod, newdata))ggplot(sleep2, aes(x = days_deprived, y = Reaction)) + geom_line(data = newdata2, color = '#f4cae2', size = 1.5) + geom_point() + theme_bw() + scale_x_continuous(breaks = 0:7) + facet_wrap(~Subject, nrow = 3) + labs(y = "Reaction Time", x = "Days deprived of sleep (0 = baseline)")Partial pooling
It would be desirable to improve our estimates for individual participants by taking advantage of what we know about the other participants.
It would improve generalization to the population.
We are interested in the 18 subjects as examples drawn from a larger population of potential subjects.
Partial pooling
When partial-pooling goes wrong...
Vanishing confidence intervals (the model is too complex for the data to support).
Unintended Shrinkage.1
- In 2016, George et al. showed that partial pooling can lead to artificially low predicted mortality rates for smaller hospitals.
The number of random effect levels has a strong impact on the uncertainty of the estimate variance.2
The estimates are pulled or "shrunk" closer to the overall population average.
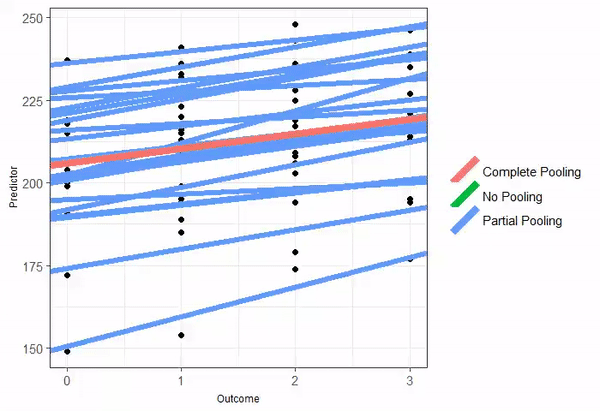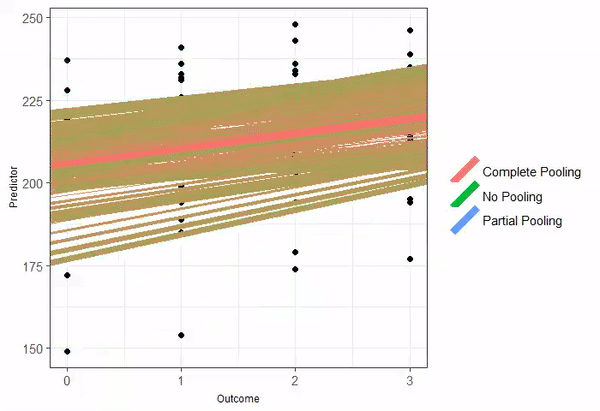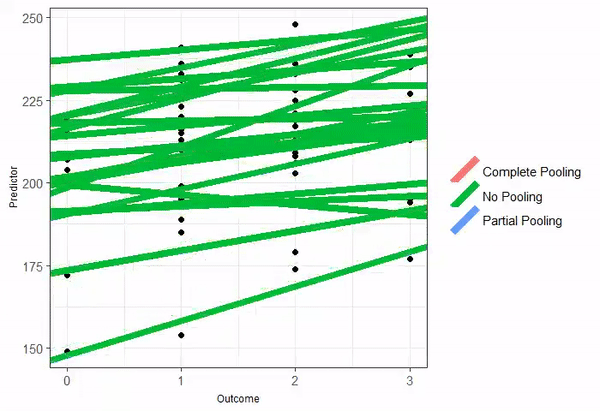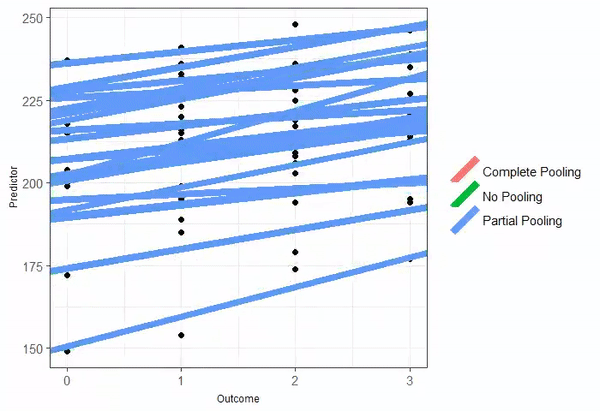 The animated GIF is from Isabella R. Ghement. Twitter: @IsabellaGhement
The animated GIF is from Isabella R. Ghement. Twitter: @IsabellaGhement
1 https://towardsdatascience.com/when-mixed-effects-hierarchical-models-fail-pooling-and-uncertainty-77e667823ae8 2 https://www.muscardinus.be/2018/09/number-random-effect-levels/
Report writing
Goal: Reproducible
Describe and justify your model (summarize the model fit).
- Specify all fixed and random effects, interactions, intercepts, and slopes
- Specify sample size: number of observations, and number of levels for each grouping factor
Mention that you checked assumptions, and that the assumptions are satisfied.
Fixed effects: Report significance (test statistic, degrees of freedom, p-value) and actual estimates for coefficients (effect size: magnitude and direction, standard errors, and confidence intervals). Report Post-hoc tests (
emmeans()).Random effects: Report the variances, standard deviations, and any correlations for each component
Cite the relevant packages.
(recommended) Basic descriptive statistics (e.g. mean and standard deviation of each group)
(recommended) Plot showing distribution of the data
Post-hoc tests test for differences between (combinations of) levels of a factor x, after fitting the model.
Why LME model?
Multilevel / Hierarchical / Nested data
Within-subject factor? Repeated measures?
Psedoreplications (multiple measurements within the same condition)?
Multiple stimulus items?
Science (causal) model before statistical model
Data types: numerical continuous data or categorical data?
Operational hypothesis?
In the sleepstudy data, you had the within-subject factor of Day (which is more a numeric variable, actually, than a factor; but it has multiple values varying within each participant).
generalized linear model (GLM) η determines the predicted mean μ of Y (link function) The predicted mean is just the linear predictor (linear regression)
take a step back (McElreath, 2022)
References
Winter, B. (2013). Linear models and linear mixed effects models in R with linguistic applications. arXiv:1308.5499. [http://arxiv.org/pdf/1308.5499.pdf]
Shravan Vasishth, Daniel Schad, Audrey Bürki, Reinhold Kliegl. (on going) Linear Mixed Models in Linguistics and Psychology: A Comprehensive Introduction. [https://vasishth.github.io/Freq_CogSci/]
Barr, Dale J. (2021). Learning statistical models through simulation in R: An interactive textbook. Version 1.0.0. Retrieved from https://psyteachr.github.io/stat-models-v1.
McElreath, R. (2020). Statistical Rethinking: A Bayesian course with examples in R and STAN. CRC Press.
Further Readings:
Casella, George, and Roger L Berger. 2002. Statistical Inference. Second Edition. Cengage Learning.
Miller, I., and M. Miller. 2004. John e. Freund’s Mathematical Statistics with Applications. Prentice Hall.
Barr, Dale J, Roger Levy, Christoph Scheepers, and Harry J Tily. 2013. “Random Effects Structure for Confirmatory Hypothesis Testing: Keep It Maximal.” Journal of Memory and Language 68 (3): 255–78.
Bates, Douglas M., Reinhold Kliegl, Shravan Vasishth, and Harald Baayen. 2015. “Parsimonious Mixed Models.”
Thanks!

Slides created via the R package xaringan.
The chakra comes from remark.js, knitr, and R Markdown.
